Over the past two centuries, there has been a gradual paradigm change in the way we look at chronic inflammatory airway diseases, including asthma and chronic obstructive pulmonary disease (COPD), shifting away from prototypic, organ-centred disease labels, coined by William Osler at the end of the 19th century, to differentiated disease subsets [1]. During the second half of the past century, asthma has been mostly perceived as an allergen-driven, T-helper-2 disorder with mast cells and eosinophils as key effector cells. This concept was based on the hygiene hypothesis and reinforced by experimental studies in mice. Consequently, all new asthma medications were tested in the allergen challenge model: given its responsiveness to corticosteroids and an overall good predictive value (albeit based on a selected population), this resulted in a one-size-fits-all approach reflected by concurrent guidelines during 1980s-2000, advocating a stepped-treatment approach. After an initial improvement in asthma outcomes, increasing evidence showed that (high-dosed) corticosteroids may cause clinically relevant adverse effects [2], especially when given systemically, and that despite optimal controller treatment and adequate adherence, still too many patients remain sub-optimally controlled [3,4]. Clearly, this called for a differentiated approach to disease management and treatment.
In the past two decades, innovative tissue sampling methods combined with omics-technologies and systems biology approaches expansively increased our understanding and insights into the mechanisms and molecular pathways underlying several chronic inflammatory diseases [5,6]. In the context of asthma and COPD, novel insights have revealed a vast complexity (i.e. consisting of several components) and heterogeneity (i.e. indicating that not all components are present within a given patient over time) within and across disease subsets, especially in severe asthma [7,8]. As a next step, clinical/inflammatory phenotypes and endotypes have been defined [7]. A ‘phenotype’ reflects an individual’s observable characteristics, including clinical, imaging, functional, physiological, and biological features, while ‘endotype’ refers to the cellular and molecular pathways underlying the pathogenesis of the disease [9].
These novel views drove the concept of precision medicine for asthma [10]. In 2016, a panel of experts proposed a precision medicine strategy for chronic inflammatory airway disease based on the presence of so-called “treatable traits” [9]. This pragmatic approach advocates active identification, assessment, and treatment of (label-free) treatable traits, consisting of pulmonary, extra-pulmonary and behavioural, psychological or lifestyle-related treatable characteristics and risk factors. The goal of this approach is to link the right patient to the right treatment and, thus, to improve clinical outcomes for individual patients and to minimise unnecessary side effects in non-responders. In this context, validated biomarkers, such as the amount of eosinophils in peripheral blood or sputum and the fraction of exhaled nitric oxide (FeNO), can serve as indicators of a specific phenotype or endotype and can help to identify responders to (targeted) therapeutic options [11]. Unsurprisingly, several original research and other work presented at the ATS 2019, organised in Dallas, USA, reflected or linked to the concept of treatable traits. Moreover, some of the treatable traits have also been implemented in the updated GINA guidelines [12].
Application of treatable traits
One of the first times the proposed concept of treatable traits has been applied to patient data, was within the Unbiased BIOmarkers in PREDiction of respiratory disease outcomes (U-BIOPRED) project. In total, 23 treatable traits were identified in the U-BIOPRED mild/moderate and severe asthma cohorts [13,14]. The most prevalent extra‐pulmonary traits comprised atopy, rhinosinusitis, nasal polyps, obesity, gastroesophageal reflux disease (GERD), and obstructive sleep apnoea. The prevalence of treatable traits was associated with poor clinical outcome and was generally higher in individuals with severe asthma compared to mild/moderate asthma.
At the ATS 2019, several presentations showed examples of existing and potential treatable traits, tools, and methods to assess and improve disease awareness, treatment adherence, disease control, and health-related quality of life (e.g. patient reported outcomes, self-assessments, and eHealth tools) [e.g. 15-17]. Additionally, several studies underscored the effect of (treatment of) comorbidities and risk factors on asthma severity and control, while other studies showed novel biomarkers or favourable outcomes based on biomarker-guided treatment strategies. Below, a selection of clinically impactful or thought-generating presentations.
Wojcik and colleagues compared allergen sensitisation with seasonal and perennial allergens between adult (>18 years) and paediatric (<18 years) patients with severe early-onset asthma (EOA; i.e. asthma diagnosis at <12 years) from the Severe Asthma Research Program (SARP). In contrast with children with EOA, they found that adults were less likely to have specific IgEs for seasonal allergens (grasses, trees, and weed mix). There were no differences in perennial allergens (cats, dogs, rodents, dust mites, and cockroaches) between the two age groups, while children with severe EOA were more sensitive to moulds (Alternaria, Aspergillus, and Cladosporium) than adult EOA (all P<0.004). The combination of loss of seasonal allergen sensitivity with age with persisting responsiveness to indoor allergens suggests different mechanisms in allergen sensitivity at older age. Unravelling these differences may reveal new immunologic pathways yielding potential targeted interventions [18]. Ghincea and colleagues aimed to identify unique predictors of asthma exacerbations in the obese asthma (BMI>30) cluster (n=461) of the SARP, which was defined as a distinct phenotype comprised of predominantly obese women with adult-onset, non-atopic, non-type2 (predominantly) severe asthma [19]. They found that GERD had a stronger association with asthma exacerbation (defined as emergency or urgent care visit) in obese vs non-obese asthmatics. Indeed, both obesity and GERD have been identified as treatable traits in the position paper [13] as well as in the concurrent guidelines [12]. At ATS 2019, an entire oral session, chaired by expert Prof. Anne Dixon (University of Vermont, USA), was dedicated to obese asthma, highlighting its pathophysiology, including oxidative stress, and the relationship with obstructive sleep apnoea.
Chronic rhinosinusitis with nasal polyps (CRSwNP) and type2 (severe) asthma share the same immune/inflammatory pathways and are frequent comorbidities. Positioning of targeted therapy with biologics for patients with CRSwNP with or without comorbid asthma is currently subject to expert discussions [20] and has also been addressed during the ATS 2019. Laidlaw and colleagues evaluated the efficacy of a monoclonal antibody blocking the interleukin(IL)-4 and IL-13 pathway, dupilumab, vs placebo on upper and lower airway outcome measures in a pooled patient population of two large phase 3 studies (SINUS-24 and SINUS-52) after 24 weeks of treatment on top of mometasone furoate nasal spray (MFNS) maintenance therapy. Dupilumab as an add-on to MFNS improved both upper and lower airway outcome measures vs MFNS alone in patients with severe CRSwNP and comorbid asthma after 24 weeks in the pooled SINUS-24 and SINUS-52 population and was well tolerated [21].
Apart from eosinophils and FeNO as validated point-of-care biomarkers of type2 asthma, novel and more sophisticated non-invasive methods, such as molecular profiling of exhaled breath (breathomics), are currently under development as prognostic tools [22]. The eNose has been shown to be able to differentiate between several aspects of disease vs healthy status. Van Bragt and colleagues investigated the potential of breathomics by eNose to serve as a composite biomarker to identify responders to targeted therapy, such as anti-IL-5 treatment with mepolizumab. Applying exhaled breath analysis by eNose, the researchers could discriminate between severe asthma patients who continued mepolizumab treatment (after 12 months) and those who did not (ROC-AUC: 0.81) [23].
While there is consensus that (oral) corticosteroid load should be limited, especially in those at risk for associated side effects, systemic corticosteroids are still the mainstay for treatment of exacerbations in patients with inflammatory airway disease (i.e. asthma and COPD). Sivapalan and colleagues investigated if an algorithm based on a biomarker, i.e. blood eosinophils (eosinophil-guided treatment, EGT), could safely allow oral corticosteroid reduction in hospitalised patients with acute COPD exacerbation vs standard treatment according to international guidelines (control group) [24]. A total of 318 COPD patients were randomly assigned to either EGT or standard treatment regimen within 24 hours of admission. For the EGT group, prednisolone was administered during the first 5 days only on days where the eosinophil count was ≥0.3 x 109 cells/L; on all other days, the oral corticosteroid was not administered, while in the control group, patients received prednisolone according to present guidelines (5 days). As compared with standard treatment, the biomarker approach (EGT) substantially reduced the duration (mean 2.4 vs 4.9 days; P<0.0001) and total dose (mean 261 vs 421 mg; P=0.0002) of oral corticosteroid in hospitalised COPD patients with acute exacerbation.
Treatable mechanisms underlying treatable traits
The U-BIOPRED consortium initially applied the systems biology approach to identify immune and inflammatory pathways and molecular mechanisms underlying different asthma phenotypes [25]. Validation of these pathways helped to define distinct endotypes and treatable mechanisms. Precision medicine approaches aim to link these treatable mechanisms to treatable traits and distinctive biomarkers based on clinical, physiological, and inflammatory characteristics of clinical phenotypes. A future step requires the application of bioinformatics (including machine learning techniques) enabling rapid detection of molecular mechanisms with subsequent links to targeted therapeutic options for individual patients [26].
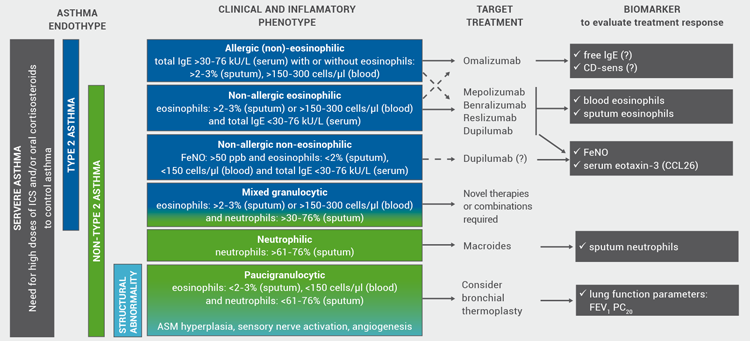
Is precision medicine still a long way to the clinic? So far, two different endotypes have been identified in severe asthma: the type2-high endotype (associated with an allergic and an eosinophilic phenotype, partly overlapping, derived from the T-helper 2 and innate lymphoid cells type 2 immune pathways) and the type2-low endotype (neutrophilic, paucigranulocytic, airway smooth muscle predominant; Figure 1), while coexistence of both endotypes has also been reported [7,27]. Presently, type2-high is the best characterised endotype and several type2-targeted treatment options have been developed. Consequently, targeted treatment options (biologics) and discriminating biomarkers are being integrated into the guidelines [12]. Targeted treatments block specific inflammatory pathways or single mediators. Single and composite biomarkers help to identify patients who will benefit from these treatments. So far, only a few point-of-care inflammatory biomarkers have been validated for clinical application [11].
In contrast, for type2-low asthma, sensitive biomarkers and targeted treatment options are largely lacking, although recent advances with omics technology showed two transcriptome-associated clusters (TAC2 and TAC3). TAC2 was associated with the highest sputum neutrophil levels, serum C-reactive protein levels, and eczema, while TAC3 showed normal to moderately increased sputum eosinophils and a higher forced expiratory volume in 1 second (FEV1) [28]. This report will not specifically discuss non-type2 treatment options.
In our recently published EAACI position paper, our task force aimed to define clinically applicable biomarkers reflecting the immune/inflammatory pathways underlying the severe asthma phenotypes and to establish the links to existing and upcoming (targeted) treatment options (Figure 1) [11].
Figure 1: Immune/inflammatory pathways underlying (severe) asthma including validated biomarkers and links to novel and existing (targeted) treatment options [11]
Reprinted from Diamant Z, et al. Allergy. 2019 [Epub ahead of print]. Copyright 2019, with permission from Wiley.
Targeted treatment options and algorithms based on precision medicine
In the early 2000s, two types of targeted therapies became available for treatment of mainly allergic asthma patients: montelukast, a small molecule leukotriene receptor antagonist (preceded by the less potent pranlukast, marketed in Japan) and omalizumab, a biologic binding free serum IgE. Since the past decade, several novel biologics have entered the clinic: all targeting the eosinophil/type 2 inflammatory pathways, while many others are currently in several stages of drug development with both upstream targets (e.g. the alarmins: TSLP, IL-33, and IL-25) and (more) downstream targets (e.g. IL-17, CRTH2/DP2, MMPs, GATA3, ST2, IL-6, PI3K) [11,29]. Currently available and some of the emerging targeted therapies are summarised in Table 1.
Table 1: Currently available and emerging targeted therapies for chronic inflammatory (airway) disease
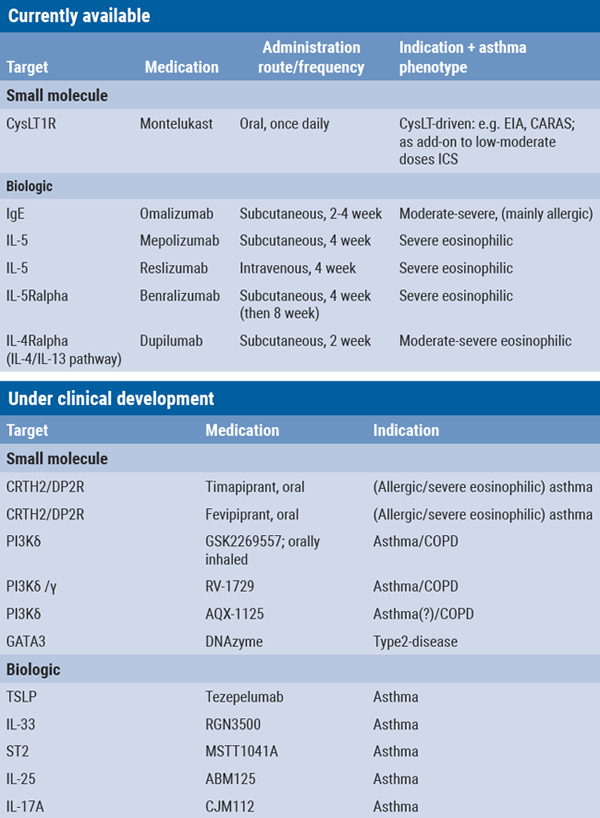
CysLT1R, cysteinyl leukotriene receptor; EIA, exercise-induced asthma; CARAS: concomitant allergic rhinitis and asthma syndrome; CRTH2, chemoattractant receptor-homologous molecule expressed on T-helper 2 cells; DP2R, prostaglandin D2 receptor; ICS, inhaled corticosteroid; PI3K, phosphoinoside-3-kinase; TSLP, thymic stromal lymphopoietin; IL, interleukin; ST2 (receptor for IL-33), also known as IL-1 receptor like 1 (IL-1RL1).
At the ATS 2019, a large number of studies on existing, novel, and emerging (targeted) therapies have been presented. In addition to novel biologics, several research groups presented promising data of different triple therapies (consisting of inhaled corticosteroid, long-acting β2-agonist, and long-acting muscarinic antagonist) on clinical outcome in patients with moderate-severe asthma [30,31].
In the context of targeted therapy, several data of the recently marketed dupilumab coming from the LIBERTY ASTHMA QUEST and VENTURE phase 3 studies in moderate-severe and severe oral corticosteroid-dependent asthma, respectively, have been highlighted in an interesting poster discussion session. In these studies, dupilumab effectively improved symptoms, quality of life, lung function, and asthma control, while allowing oral corticosteroid withdrawal [32-35]. In one of the studies, eosinophilia was found in patients while on dupilumab [34], which deserves further evaluation.
To aid precision medicine based on the underlying immune/inflammatory pathways and available biomarkers, a number of treatment algorithms for uncontrolled severe asthma have been proposed [e.g. 36,37]. Based on recent developments, linking inflammatory phenotypes, validated point-of-care biomarkers, and novel targeted treatment options, our EAACI task force also proposed a treatment algorithm (Figure 2) [11]. Further insights into the highly complex, heterogeneous, and variable nature of chronic inflammatory airway disease, identification of reliable and sensitive biomarkers, as well as long-term trials with innovative therapies will further refine clinically applicable treatment algorithms for severe asthma.
Figure 2: Treatment algorithms for severe asthma linking inflammatory phenotypes, biomarkers and targeted treatment options. Modified from Diamant et al. (2019).
Conclusions, questions, and unmet needs
So far, biologics have shown a substantial improvement in disease outcome and allowed reduction of corticosteroids in many patients with severe asthma with eosinophilic airway inflammation. However, not all patients respond equally well, despite similar clinical, functional, and inflammatory characteristics; and in some patients, response may vary over time. Furthermore, dissociation in inflammatory vs clinical parameters has been reported in response to anti-IL-5 treatments in patients with ‘eosinophilic’ COPD (sputum eosinophils >3%) [38]. In this pilot study, add-on mepolizumab for 6 months failed to show efficacy on lung function parameters, exacerbation rate, or quality of life in smoking COPD patients, despite a substantial reduction in sputum and blood eosinophils. Similar data were found in the GALATHEA and TERRANOVA trials, where add-on benralizumab for 1 year failed to reduce exacerbation rates in frequently exacerbating patients with moderate to very severe eosinophilic COPD (blood eosinophils ≥220 cells/mm3) [39]. This heterogeneity in treatment response may be caused by differences in molecular pathways or endotypes underlying eosinophilic airway inflammation, different co-existing immune/inflammatory signatures in individual patients, external factors/triggers, and/or the effect of adaptive and innate immune responses in different subsets. These observations obviously call for further research.
Other questions to answer
Given the complex, variable, and heterogeneous nature of the molecular pathways underlying difficult to control, severe chronic inflammatory airway disease:
- Will targeting one single pathway treat all aspects of the disease and, thus, be sufficient to achieve “ultimate” control?
- Will combination therapy (e.g. ≥1 biologics, small molecules, or combinations) be safe and help to maintain clinical effectiveness over time?
- Are targeted therapies disease-modifying?
- Are pathways underlying biologics response different in paediatric asthma patients compared to adult patients?
Unmet needs
- Long-term efficacy trials (consistency, variability, heterogeneity of treatment response, and biomarkers).
- Further identification of molecular pathways underlying severe chronic inflammatory airway disease.
- Reliable (composite) biomarkers predicting and monitoring treatment response.
- Less expensive production methods of biologics.
- Diamant Z, Diderik Boot J, Christian Virchow J. Respir Med. 2007;101(3):378-88.
- Price DB, et al. J Asthma Allergy. 2018;11:193-204.
- Rabe KF, et al. Eur Respir J. 2000;16(5):802-7.
- Bateman ED, et al. Am J Respir Crit Care Med. 2004;170(8):836-44.
- Thamrin C, et al. Am J Respir Crit Care Med. 2016;194(9):1053-61.
- Silkoff PE, Moore WC, Sterk PJ. Clin Chest Med. 2019;40(1):13-28.
- Wenzel SE. Nat Med. 2012;18(5):716-25.
- Sakornsakolpat P, et al. Nat Genet. 2019;51(3):494-505.
- Agusti A, et al. Eur Respir J. 2016;47(2):410-9.
- Jameson JL, Longo DL. N Engl J Med. 2015;372(23):2229-34.
- Diamant Z, et al. Allergy. 2019. doi: 10.1111/all.13806. [Epub ahead of print]
- Global Initiative for Asthma. Accessed at [www.ginasthma.org](https://ginasthma.org/).
- Agusti A, et al. Eur Respir J. 2017;50(4).
- Simpson AJ, et al. Allergy. 2019;74(2):406-11.
- Ali A, et al. Am J Respir Crit Care Med. 2019;199:A3014.
- Jones R, et al. Am J Respir Crit Care Med. 2019;199:A3028.
- Koltowski L, et al. Am J Respir Crit Care Med. 2019;199:A3036.
- Wojcik R, et al. Am J Respir Crit Care Med. 2019;199:A4076.
- Ghincea A, et al. Am J Respir Crit Care Med. 2019;199:A1282.
- Fokkens WJ, et al. Allergy. 2019.
- Laidlaw T, et al. Am J Respir Crit Care Med. 2019;199:A7356.
- Boots AW, et al. Trends Mol Med. 2015;21(10):633-44.
- Van Bragt JMH, et al. Am J Respir Crit Care Med. 2019;199:A2673.
- Sivapalan P, et al. Am J Respir Crit Care Med. 2019;199:A7352.
- Kaminsky DA, Irvin CG, Sterk PJ. J Appl Physiol (1985). 2011;110(6):1716-22.
- Chung KF, Adcock IM. Allergy. 2019. doi: 10.1111/all.13771. [Epub ahead of print]
- Erjefalt JS. Curr Opin Pulm Med. 2019;25(1):79-86.
- Kuo CS, et al. Eur Respir J. 2017;49(2).
- Corren J. J Allergy Clin Immunol Pract. 2019;7(5):1394-403.
- Paggiaro P, et al. Am J Respir Crit Care Med. 2019;199:A7080.
- Watz H, et al. Am J Respir Crit Care Med. 2019;199:A7081.
- Hanania NA, et al. Am J Respir Crit Care Med. 2019;199:A2667.
- Rabe KF, et al. Am J Respir Crit Care Med. 2019;199:A2668.
- Sher L, et al. Am J Respir Crit Care Med. 2019;199:A2669.
- Ford B, et al. Am J Respir Crit Care Med. 2019;199:A2666.
- Svenningsen S, Nair P. Front Med (Lausanne). 2017;4:158.
- Blaiss MS, et al. Ann Allergy Asthma Immunol. 2017;119(6):533-40.
- Dasgupta A, et al. Eur Respir J. 2017;49(3).
- Criner GJ, et al. N Engl J Med. 2019. doi: 10.1056/NEJMoa1905248. [Epub ahead of print]
Posted on
Previous Article
« Aclidinium bromide delays COPD exacerbation without increased MACE risk Next Article
SAATELLITE trial: Suvratoxumab prevents ventilator-associated Staphylococcus Aureus pneumonia in intensive care unit patients »
« Aclidinium bromide delays COPD exacerbation without increased MACE risk Next Article
SAATELLITE trial: Suvratoxumab prevents ventilator-associated Staphylococcus Aureus pneumonia in intensive care unit patients »
Table of Contents: ATS 2019
Featured articles
Letter from the Editor
Interview with Prof. Christian Bergmann
Treatable Traits in Chronic Inflammatory Airway Disease: Back to Basics
Treatable traits in chronic inflammatory airway disease: back to basics
Critical Care Medicine
Distinguishing between 4 different subtypes of sepsis sets the stage for individualised treatment
Stem cell therapy in acute respiratory distress syndrome improves 28-day mortality
SPICE III trial: Early sedation with dexmedetomidine in critically ill patients
SAATELLITE trial: Suvratoxumab prevents ventilator-associated Staphylococcus Aureus pneumonia in intensive care unit patients
Sleep Medicine
Million-patient study reveals gaps in long-term adherence among various sub-populations
Sleep apnoea severity has a non-linear relationship with acute myocardial infarction risk
Obstructive sleep apnoea affects morning spatial navigational memory processing in asymptomatic older individuals
Pulmonary Vascular Disease and Interstitial Lung Disease
Nintedanib reduces lung function decline in systemic sclerosis-associated ILD
Pulmonary arterial hypertension: early treatment with selexipag most effective
Long-term safety and efficacy of recombinant human pentraxin-2 in patients with idiopathic pulmonary fibrosis
Infection
Dupilumab improves outcomes in patients with severe chronic rhinosinusitis with nasal polyps and comorbid asthma
Durability of culture conversion in patients receiving ALIS for treatment-refractory MAC lung disease
E-cigarette use disrupts normal immune response to viral infections, particularly in women
Paediatric Pulmonary Medicine
Bacterial pneumonia predicts ongoing lung problems in infants hospitalised for acute respiratory failure
Aspergillus and early cystic fibrosis lung disease: does it need to be treated?
COPD
CORTICO-COP trial: eosinophil-guided therapy reduces systemic corticosteroid exposure
A randomised controlled trial of a smoking cessation smartphone application
Benralizumab does not ameliorate COPD exacerbations (GALATHEA/TERRANOVA trials)
Aclidinium bromide delays COPD exacerbation without increased MACE risk
Bench-to-Bedside (Pre-Clinical)
Human lung organoids to study foetal RSV infection
CRISPR/Cas9 genome editing therapy of hereditary pulmonary alveolar proteinosis
Cilia diagnostics in primary ciliary dyskinesia
Tuberous sclerosis complex 2 may be a novel target in pulmonary arterial hypertension therapy
Related Articles
November 28, 2019
Smart shirt as a device to measure tidal volumes in real-life setting
© 2024 Medicom Medical Publishers. All rights reserved. Terms and Conditions | Privacy Policy

